
After a couple of months from the 3DBIONOTES-COVID19 tool first release, there has been made a new update to the dedicated website to collect all available SARS-CoV-2 virus structural data: https://3dbionotes.cnb.csic.es/ws/covid19
The Spanish National Centre for Biotechnology (CNB-CSIC), part of the INB/ELIXIR-ES, is now presenting an upgrade with very interesting new features. They have made a huge effort to improve performance, consolidate data and refactor code, although not all changes are visually evident from the outside.
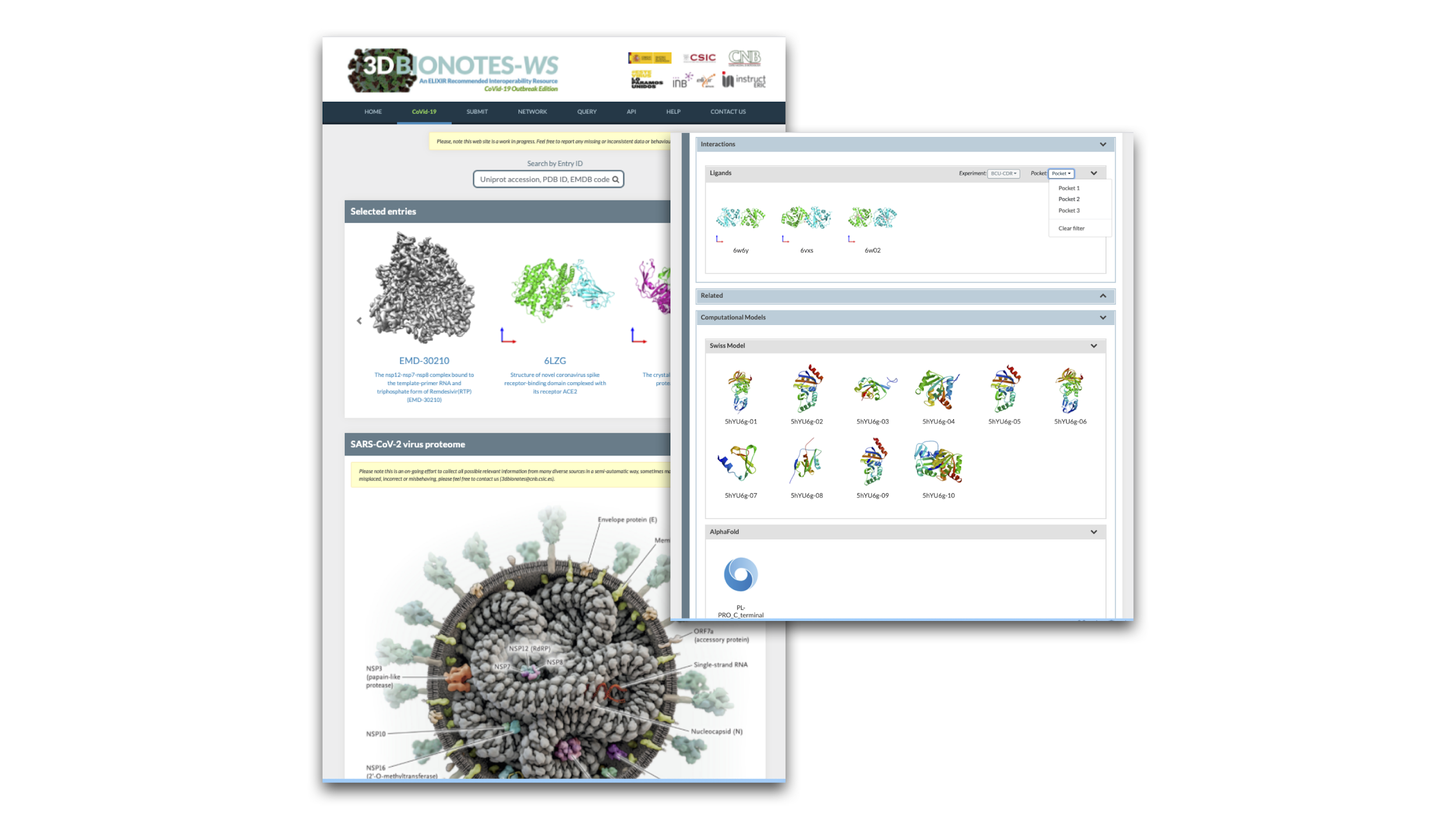
New Features
- New hyper realistic infographics representing a virus used as a map index. Users can click on a protein label and will be directed to the corresponding section.
- Added computational models from various sources: SwissModel, AlphaFold & BSM-Arc. These are especially useful for those proteins with missing experimental models.
- New Ligand interactions section including results from experimental and computational drug screening & repurposing, one of them from our group to be published in short.
- New filters for Ligand interactions enabling selection by experiment and interaction pocket.
Updates
- Improved loading for validated models from PDB-Redo and Isolde.
- Updated help and documentation.
The 3DBIONOTES Application Programming Interface (API) is an ELIXIR Recommended Interoperability Resource. This reusable platform-independent is key to gene metadata alignment, annotation, and integration across major bioinformatics data resources.
The Biocomputing Unit of the CNB-CSIC in Madrid hosts the 3DBIONOTES-COVID19 under the leadership of Prof José María Carazo and Dr Carlos Óscar Sánchez Sorzano, and in cooperation with INB/ELIXIR-ES and Instruct-ERIC (Instruct Image Processing Centre).
